
Arbregen

 |
Arbregen |
 |
|
An overview of the software"Arbregen" You'll find more substantial explanations in the help file that comes with the software. |
Arbregen allows you to build pedigrees and then compute the odds of genotypes for the individuals in the tree.
The trait is supposed to result of the expression of one gene with two possible alleles.
The transmission mode may be either autosomic or sex-linked, the alleles may be dominant or recessive.
The exact computation is possible but quickly becomes tedious as the tree reaches some complexity.
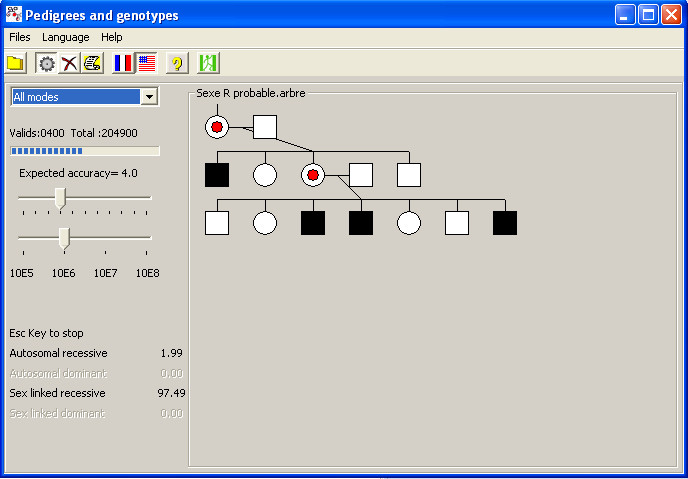
No premises are added about genotypes, beyond the ones built into the pedigree when designed by the user.
Those include phenotypes, genders, and assumptions about individuals at the borders of the tree (for example the probability for the root parents to be heterozygous). The software starts by giving a genotype to the root parents. It does this randomly, and according to the chosen a priori odds. It then mimics the meioses by picking an allele from each parent to provide each child with a genotype. If any incompatibility appears between the elected genotype and the actual phenotype built in the tree, the whole tree is discarded (for example, if one of the parents receives a homozygous normal genotype and one of their children shows the recessive phenotype). Only the trees which are fully data compatible will make it through.
Sorting out the odds of each genotype for each individual is easy at this point.
For instance 10000 trees have been computed, and 1000 appear not to be self-contradictory.
If 600 trees show the II-2 individual as being heterozygous and the other 400 as homozygous,
then the odds for that individual to be heterozygous are 6 to 4 (i.e. a 60%probability).
The whole process is iterated, using different transmission modes
(the software allows the user to restrict the computation to one or several particular modes).